This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Gene Phylogeny
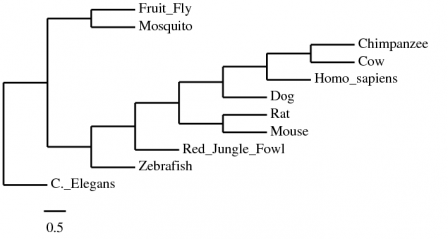
Phylogeny.fr is a program that allows one to determine phylogentic relationships between homologous genes and proteins. Using the MUSCLE alignment, TNT phylogeny, and TreeDyn tree rendering features, the following phylogenetic tree was established (Figure 1) among the DNA sequences of the different homologs of parkin. The advantage of this tree is that it has parsimony; that is, the phylogeny was constructed using the fewest evolutionary events possible, which lends credibilty to its likelihood. The disadvantage of this tree is that it does not contain any branch support values.
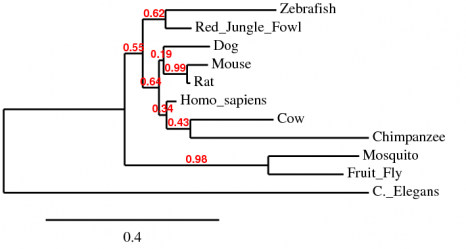
Using 3D Coffee alignment, PhyML phylogeny,and TreeDyn tree rendering, the following phylogenetic tree was established (Figure 2) along with corresponding branch support values. It is noteworthy that monophyletic groups between the two trees are significantly different, specifically with respect to the separation of vertebrates and invertebrates. Because the monophyletic groups of the tree below more closely follow the homolog alignments, my preference is to use this tree:
References
1. Phylogeny.fr. www.phylogeny.fr